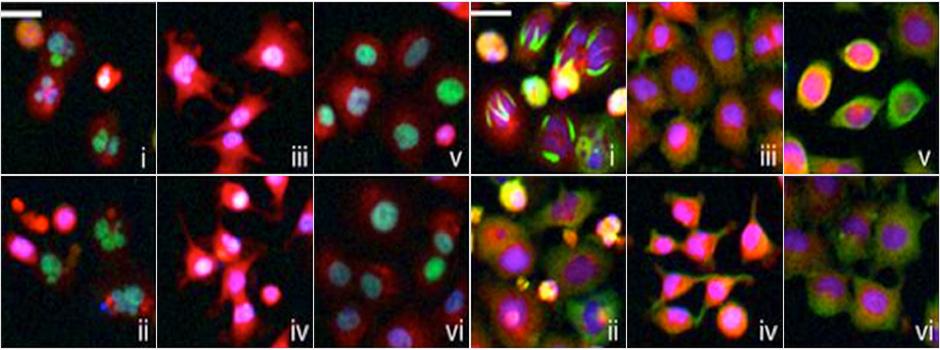
The search for a cure to cancer is the focus of so many efforts, including the pursuit for even more effective tools to expedite that discovery. Screening cancer drugs is complicated and colorful, with time consuming, conventional methods requiring many screenings before getting meaningful results. A recent article in Nature Biotechnology proposes a novel concept to enhance how scientists look at cells--with high-content image analysis and drug screening to interpret the different stresses cells undergo and predict the types of drugs that will cure them.
NYU Shanghai Professor of Biology Jungseog Kang is among several authors who have found a means of identifying lead compounds across multiple drug classes at a single-pass screen, e.g. "high-content image-based phenotypic screening.” While most cancer drug screenings aim to find one class of drug compounds in a single screen, the new method, high-content screening, maps out several different classes of leading drug compounds in a single screen.
For screening, cancer cells are exposed to different drug types and their response to those drugs show which part of the cell has been targeted. A common painstaking screening method only identifies one target at a time. The larger the drug compound library, the greater number of shots/scans are needed to effectively measure all possible targets.
High content screening identifies different types of drugs in a single-pass screen while specificity is still assured. The researchers demonstrated, in a single-pass screen, that a single “reporter” cell line accurately identified lead compounds simultaneously across a range of drug classes.
In a non-biased and systematic way, the study generated a library of reporter cell lines (like a library of genes, each having a unique function). It shows different fluorescent tagged proteins while locating the ones for the best screening results, and a tagged gene is visualized as fluorescent when it has been translated into a protein. Using analytical criteria, the authors identified a "reporter" protein in this library, one that accurately classifies drugs across multiple classes.
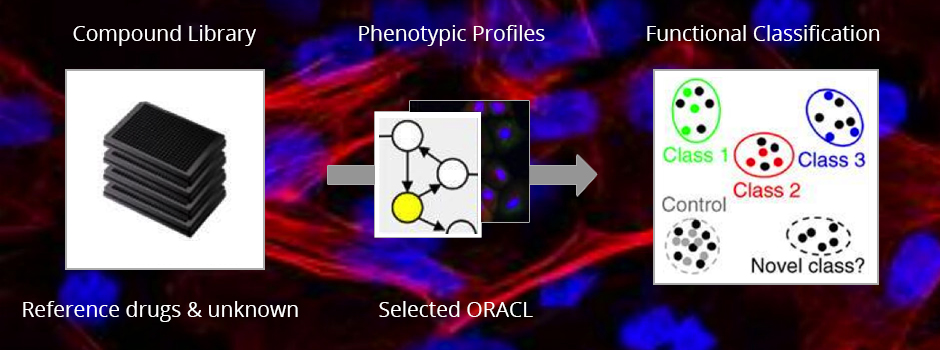
The study demonstrated an effective, new method of selecting the optimal reporter protein in a systematic way. In effect, an “informative reporter” cell line in a high-content drug screen can be systematically selected to increase its efficiency, scale, and accuracy by maximizing its discriminatory power. This new method provides more information from cellular samples and answers more questions simultaneously with information-rich data sets--producing meaningful results and a more productive lab.
The study’s other authors include: Chien-Hsiang Hsu (UCSF), Qi Wu (UTSW), Shanshan Liu (UTSW), Adam D Coster (UTSW), Bruce A Posner (UTSW), Steven J Altschuler (UCSF) and Lani F Wu (UCSF).